P-sort: an open-source software for cerebellar neurophysiology
Analysis of electrophysiological data from Purkinje cells (P-cells) of the cerebellum presents challenges for spike detection. Complex spikes have waveforms that vary significantly from one event to the next, raising the problem of misidentification.
Abstract
Analysis of electrophysiological data from Purkinje cells (P-cells) of the cerebellum presents challenges for spike detection. Complex spikes have waveforms that vary significantly from one event to the next, raising the problem of misidentification. Even when complex spikes are detected correctly, the simple spikes may belong to a different P-cell, raising the danger of misattribution. Here, we analyzed data from over 300 P-cells in marmosets, macaques, and mice, using an open-source, semi-automated software called P-sort that addresses the spike identification and attribution problems. Like other sorting software, P-sort relies on nonlinear dimensionality reduction to cluster spikes. However, it also uses the statistical relationship between simple and complex spikes to merge seemingly disparate clusters, or split a single cluster. In comparison with expert manual curation, occasionally P-sort identified significantly more complex spikes, as well as prevented misattribution of clusters. Three existing automatic sorters performed less well, particularly for identification of complex spikes.
Introduction
Recording neuronal activity from the cerebellum presents both opportunities and challenges. The principal cells of the cerebellum, Purkinje cells (P-cells), can be identified based on their unique electrophysiological properties. Among cells in the cerebellum, only P-cells can produce simple and complex spikes (Thach, 1967). This makes it possible to use statistical methods to measure the likelihood that the recorded neuron is a P-cell: generation of a complex spike should be followed by suppression of simple spikes (Eccles et al., 1966; Sato et al., 1992). However, detection of complex spikes is difficult because these spikes are not only rare, but their waveforms also vary from one spike to the next. Thus, it is common to detect the simple spikes but not the complex spikes, or alternatively, detect the complex spikes but later realize that they are not followed with simple spike suppression and therefore do not belong to the same P-cell. To address these issues, we developed a spike analysis software that aids detection of simple and complex spikes, as well as quantifies whether the two events are generated by a single P-cell.
Unlike simple spikes, the power spectrum of complex spikes tends to be greatest in the low- frequency range (30-800 Hz). As a result, a typical complex spike can produce a “broad spike” in the low-pass filtered representation of the data (local field potential, LFP). Indeed, two recent developments in complex spike detection are novel algorithms that depend partly on the LFP waveform (Markanday et al., 2020; Zur & Joshua, 2019). Once the simple and complex spikes are labeled, the final step is to determine whether the simple spikes have been suppressed after a complex spike. If so, then one may conclude that the two kinds of spikes were generated by a single P-cell. However, in some data sets complex spikes do not have an LFP signature. Moreover, even if the complex spikes are detected, the detected simple spikes may belong to a different P-cell, or even a non P-cell.
As a result, the problem is two folds: in the identification step, we need to label the simple and complex spikes, whereas in the attribution step, we need to determine which group of complex spikes was generated by the P-cell that produced a particular cluster of simple spikes. To consider these challenges, we formed a collaboration that included laboratories which focused on marmosets, macaques, and mice. Our software was developed using a database of over 300 P-cells recorded in three species.
The diversity of species and recording electrodes helped us identify some of the critical issues that are present in cerebellar electrophysiology. The presence of experts from the various laboratories provided a diversity of opinions, helping us verify the algorithms, as well as highlight their limitations. Here we report the results of this effort.
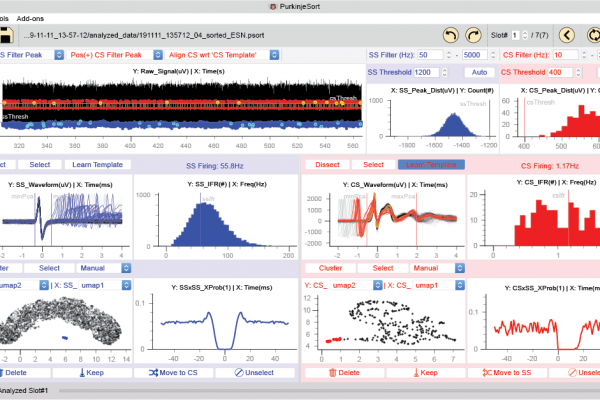
P-sort is an open-source, Python-based software that runs on Windows, MacOS, and Linux platforms. To cluster waveforms and identify simple and complex spikes, P-sort uses both a linear dimensionality reduction algorithm and a novel nonlinear algorithm called UMAP (Uniform Manifold Approximation and Projection) (McInnes et al., 2018). Importantly, it quantifies the probabilistic interaction between complex and simple spikes, providing an objective measure that can split a single cluster, or merge two different clusters, despite similarities or differences in their waveforms. Thus, P-sort helps the user go beyond waveforms to improve clustering of spikes
However, a limitation of P-sort is that it relies on user interaction. To encourage development of more automated algorithms for the cerebellum, they provide a large database of labeled spikes from all three species and P-sort’s source code available at:
https://github.com/esedaghatnejad/psort




Related Posts